Progenesis QI
Discover the significantly changing compounds in your samples
Progenesis QI Software enables you to accurately quantify and identify compounds in your samples that are significantly changing, with a highly intuitive menu-guided workflow. Progenesis QI Software helps to overcome your data analysis challenges, enabling you to rapidly, objectively, and reliably discover compounds of interest and export results for omics research applications.
Progenesis QI is instrument platform independent, with support for universal formats such as .mzML and .mzXML. For more information about Progenesis QI, see www.nonlinear.com.
Overview
- Search compound data flexibly using LipidBlast, ChemSpider, Elemental Composition, Metlin, or the Waters search method MetaScope
- Reduce false positive IDs by using multiple search parameters: exact mass, MS/MS fragments, isotope distribution, RT, and CCS
- Import identification results, including chemical structures from SDF databases and embedded web-links to compound details
- Find significantly changing compounds quickly, e.g., Anova p-value, fold-change, power
- Visualize relative compound abundance using compound abundance profile graphs
- Easily integrate results with other bioinformatics platforms with comprehensive data export options
- Review and edit the compound adduct deconvolution used to quantify and identify a compound
Recommended Use: For rapidly, objectively, and reliably discovering compounds of interest in LC-MS data analysis.
Features Header
Unlock complex data with powerful visualization
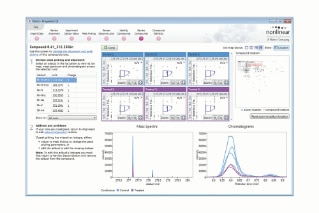
One of the key features of Progenesis QI is its highly graphical and attractive data displays which enable users to fully visualize and understand data, giving added confidence in the measurements made. Ion-intensity maps showing the 2D-representation of chromatographic retention time, m/z, and feature intensity, as well as MS and chromatographic data views, provide quality assurance of automatic alignment, peak picking, and compound adduct deconvolution for every sample.
Highly accurate quantification of compounds
Progenesis QI provides both accurate and precise measurements of compounds based on their summed ion intensities. Compound adducts are automatically deconvoluted to provide accurate quantification from all available data. Following quantification, it is possible to identify compounds having statistically significant abundance changes across groups. Large numbers of samples from multiple groups can be explored, along with multiple comparison designs within a single experiment. The result is a list of compounds of interest for further characterization.
Confident compound identification with multiple search parameters
Progenesis QI enables users to search for compound identifications based not only on neutral mass, isotopic distribution, and chromatographic retention time, but also using MS/MS fragment data and collision cross section (CCS) measurements using the Waters novel Metascope search engine. Database searching using multiple parameters greatly improves the specificity of the search and increases the chance of correct compound identification.
More easily find compounds of interest
Compound ID is still one of the main bottlenecks in small molecule omics experiments so we provide access to a wide variety of databases to increase your chances of finding your compounds of interest. Flexible compound search methods include:
- Any databases in .SDF format
- In-house databases in .CSV format
- Metlin database
- Royal Society of Chemistry databases using ChemSpider plug-in
- Lipidblast database
- NIST MS and MS/MS libraries
You can also find compounds of interest using statistical tools included in Progenesis QI, including:
- ANOVA
- Principal component analysis (PCA)
- Hierarchical clustering
You can also leverage advanced statistics via seamless integration with EZInfo 3.0 (Umetrics) for access to functions like supervised analysis and s-plots.
Reduce bottlenecks and streamline workflows
The menu-guided workflow in Progenesis QI helps guide users through the experimental steps in the software. If required, automation routines allow users to seamlessly move through multiple stages to maximize opportunities for unsupervised overnight and weekend data processing. With Progenesis QI, you’ll get:
- Automatic reference run selection and alignment
- Automatic peak picking and normalization
- Automated experimental design from .CSV and .SPL files
- All stages automatable up to the point of compound identification
Streamlining omics experiments starts with reducing the bottleneck in interpretation of the list of discoveries – what is the biology behind the changes observed? One way to address this question and extract maximum value from omics discovery data is Pathway Analysis. Progenesis QI provides export tools to quickly and easily interface with third-party Pathway Analysis programs.