Where Did My Sample Go?

A case of non-specific binding (NSB)
We’ve all been there. You spend hours (sometimes days) in the lab preparing your samples. You carefully walk them to the autosampler, lovingly load up your sample queue, and press play. Of course, you’re running these overnight because instrument time is precious and someone else needs to use it tomorrow. You come into the lab in the morning and immediately get settled in to process your data.
And there’s nothing there.
Did you forget to inject? No. Maybe you didn’t spike in your analyte at all? You’re starting to question your sanity.
“So, where did my sample go?”
If you’re confident that you haven’t made the usual mistakes (which we all do), and you know that your sample should be there, but it’s not, where did it go?
Peptides and proteins are especially prone to non-specific binding (NSB) or adsorption to solid surfaces. These surfaces could be pipette tips, sample storage containers, or injection needles. Now, we all know ways to mitigate adsorption, like:
- using carrier proteins (which can add unwanted complexity to our samples) or
- using lab supplies with high performance surfaces to help stop adsorption from happening in the first place.
However, in many cases, we may choose to go without addressing the problem.
Are you prepared to face the consequences of non-specific binding in your sample analysis?
These include:
- Low recovery
- High variability
- Poor Sensitivity
- Insufficient linear dynamic range
Lets examine these consequences further.
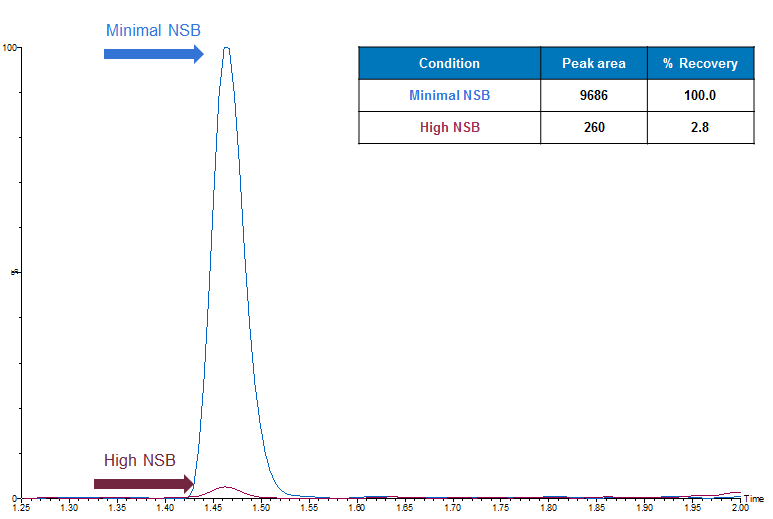
Low recovery
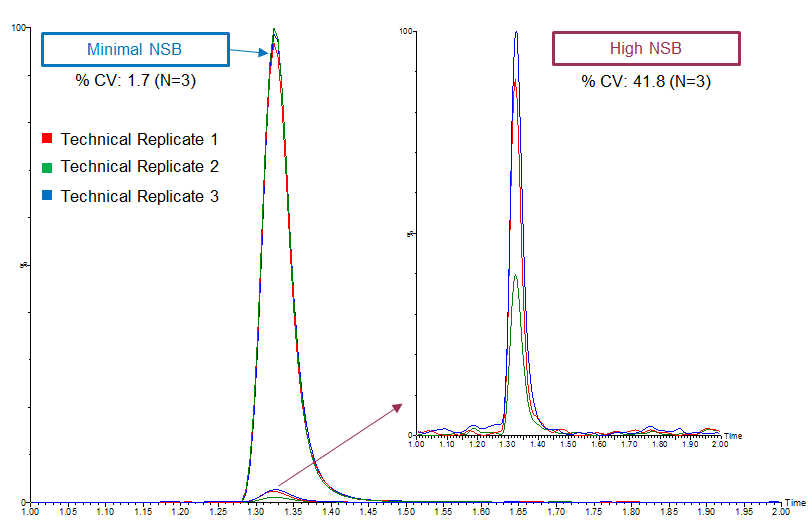
High variability
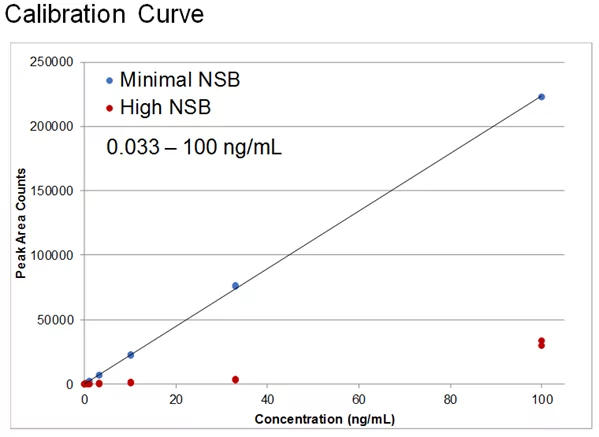
Poor sensitivity
The next time you walk into the lab and find your analyte is missing, ask yourself this:
“Did I do enough to combat non-specific binding?”
If you’re not sure, be sure to ask our scientists here at Waters for help!
Watch out for our next Blog post “Lost samples in the container: non-specific binding and the impact of blocking agents” by Moon Jung, Ph.D.
Additional resources:
Popular Topics
ACQUITY QDa (16) bioanalysis (11) biologics (14) biopharma (26) biopharmaceutical (36) biosimilars (11) biotherapeutics (16) case study (16) chromatography (14) data integrity (21) food analysis (12) HPLC (15) LC-MS (21) liquid chromatography (LC) (19) mass detection (15) mass spectrometry (MS) (54) method development (13) STEM (12)