Automating Metabolic Flux Analysis Workflows in Cancer Research
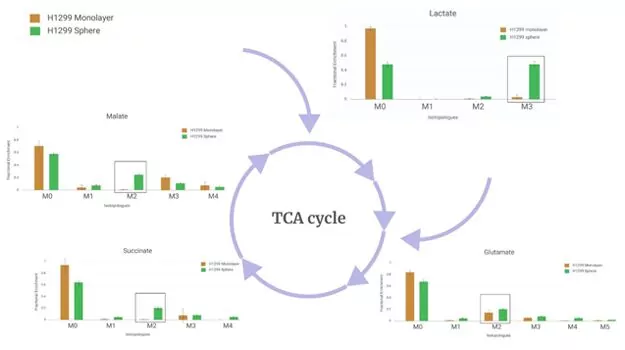
A study comparing 2D and 3D in vitro cultures in non-small-cell lung carcinoma
As a researcher focused on discovery metabolomics, one of the goals is to measure changes in a biological system’s metabolome. Based on the various metabolites identified, you can then implicate specific metabolic pathways. This form of network analysis is necessary to understand the changes diseases, such as cancer, make to a biological system.
Exactly how these intracellular pathways have changed needs to be the next question answered.
13C metabolic flux analysis (13C-MFA) has become the analytical method of choice for measuring the production and consumption rates of and consequently the dynamic changes in metabolic pathways.
As far back as 1956, Warburg et al identified the preference to aerobic glycolysis over the oxidative phosphorylation pathway in the production of energy in cancer cells. 13C-MFA has added to this knowledge implicating other altered pathways and improving our understanding of cancer metabolism.

In 13C-MFA , LC-MS data is often generated from multiple time points and conditions. These data files then need to be analyzed for the isotope incorporation in target metabolites related to the pathways of interest. This analysis requires multiple repetitive steps to be carried out on multiple data files before interpretation of the data can be undergone. Inconsistency in the processing steps can lead to errors, and waiting for one process to complete before initiating the next adds unnecessary time to the analysis.
Waters and Elucidata have worked together to remove these risks and barriers by allowing automation of the various stages of data processing. Our Symphony Data Pipeline software can now feed MSConvert and ElMaven the necessary command line arguments needed to transfer, convert, peak detect, and finally upload the experimental data files to the Elucidata Polly visualisation tools on elcudidata.io.
Working with our collaborators at Georgetown University, we put this workflow to the test with a model data set generated from a study comparing a monolayer cancer cell culture vs. a spheroid cell culture. Spheroids have been popular for some time due to their ability to better represent in-vivo micro environments. This is especially true in the case of cancer research and tumor growth. The dataset should therefore show a fairly typical representation of the pathways indicative of cancer metabolism.
You can read more about the experiment, results, and workflow in our application note.
Additional resources:
- Automating Metabolic Flux Analysis with Symphony and Polly
- 13C Qualitative Flux Analysis with Symphony Software and Polly Software of Non-Small Cell Lung Carcinoma Cells Grown in vitro in Two and Three Dimensions
For research use only. Not for use in diagnostic procedures.
Popular Topics
ACQUITY QDa (16) bioanalysis (11) biologics (14) biopharma (26) biopharmaceutical (36) biosimilars (11) biotherapeutics (16) case study (16) chromatography (14) data integrity (21) food analysis (12) HPLC (15) LC-MS (21) liquid chromatography (LC) (19) mass detection (15) mass spectrometry (MS) (54) method development (13) STEM (12)


